Does anyone with 23andme results want to “share and compare” on their website? Just got mine back.
Does anyone else have 23andMe data, which they have not shared yet (either on Solve or here)? I did some further analysis of all data previously published by PFS patients and have found all patients to have a mutation of MTRR A66G.
MTRR enzymes play a central role in the methyl group metabolic pathway, that are involved in both DNA methylation and DNA synthesis. Needless to say, these are key pathways. A recent meta study has found
that the MTRR A66G polymorphism is associated with significantly increased cancer risk
A MTRR polymorphism could contribute to our susceptibility to get PFS, while others can consume 5ARI’s without apparent problems.
This is an important information to communicate to our researchers (which I will do shortly). If you have 23andMe data which you have not previously shared, please do so and upload your results, specifically from GeneticGenie.
Thanks.
Would you recommend I get a 23andMe test done awor? I’d definitely be up for it if it would provide useful?
At this point, I am really not sure if the data is significant, and need to do further analysis. If a PFS patient had some 23andMe data lying around, I would love to increase the statistical power a little for what I am doing, but it is not worth it to get the tests done if you don’t already have them. Incidentally, I would also be very interested to get some more data from guys who don’t have PFS, or only a very mild form (again, already available data). If the scientists then want to see more, then we can take it from there.
Btw, If anyone out there has a strong background in statistics, I would love for you to give me a hand.
Hey awor,
I’m new to the site, but have been on the fence about joining for a while. I’m the admin at the PSSD Forum and run the PSSD website “PSSD Lab”. I recently saw a post about this site looking for PSSD people as well, and then today someone reposted a link to this site because I’ve actually been doing something very similar for a while on the PSSD forum.
I have a fairly strong stats background and just graduated w/ Biology and Neuroscience degrees this spring. I’d love to help this project in any way I can. I’ll go through the rest of this thread tonight. Most if not all of what I’ve been working on probably works for you all too.
What I’m trying to run is called a “Genome-Wide Association Study (GWAS)”. The numbers we have are just too low for any real analysis on the PSSD forum (Around 20). I’ve raised the money for the computer I’ll need, have spoken to researchers running these experiments, and am ready to do this if we get enough people.
We’ve written a program that runs GWAS. It works well, but it is in Python and therefore is slower than some of the programs written in C# (PLINK is the one that comes to mind).
I’ll link to the project thread and the program. I’ve locked the results for now because I’m trying to incentivize more people to send in genomes before I release.
I will say however that nearly EVERYONE (I think 90%+) has a mutation in one of their Vitamin D Receptor genes. VDR Taq and VDR Bsm are genes that PFS should look at.
http://www.pssdforum.com/viewtopic.php?f=44&t=1993
PS: There are some good places to look for control data. Open SNP project is nice. Reducing confounding variables will help if you get ppl who took fin but didn’t get PFS, but might be harder to get them to donate. Maybe posting at hairloss forums would be of use.
That’s really interesting. We do too. It’s the most frequent homozygous mutation I can see in the results, and one I personally have (VDR Taq). Thanks Ghost and welcome, glad to have you here. We’ll PM you.
Hey Ghost
First of all, a very warm welcome to you - great to see you on this site. You may recall, we had some email exchanges a while back. I was meaning to get into contact with you over the past week - fantastic that you beat me to it. Funny that you mention GWAS, I first wanted to mention it in what I am looking for, decided not to as to not scare off any potentials. And there you come along.
I have been slowly hacking away at my spreadsheet (sorry, no PLINK in use yet), looking up frequency distributions in dbsnp and so forth. Quite tired atm, but will get back to you within the next days via pm, as @axolotl wrote. I think we have some interesting and important things to discuss in the overall picture of our respective diseases.
Hi - I did 23andMe and would be happy to share my results.
Welcome to Propeciahelp Ghost! Glad to have you here.
That would be great, thanks for your offer. For the moment and for what I am doing, it would serve me well if you could just give me your GeneticGenie report, similar as to others who have posted in the past. You can either pm me or post it here, whatever you prefer. I am then hoping to look at it together with @Ghost, he might be interested in raw data by the sound of it. You can always give us that later, if required.
Gene & Variation rsID Alleles Result
COMT V158M rs4680 AG +/-
COMT H62H rs4633 CT +/-
COMT P199P rs769224 GG -/-
VDR Bsm rs1544410 CC -/-
VDR Taq rs731236 AG +/-
MAO A R297R rs6323 T +/+
ACAT1-02 rs3741049 GG -/-
MTHFR C677T rs1801133 GG -/-
MTHFR 03 P39P rs2066470 AG +/-
MTHFR A1298C rs1801131 GT +/-
MTR A2756G rs1805087 AG +/-
MTRR A66G rs1801394 AG +/-
MTRR H595Y rs10380 CT +/-
MTRR K350A rs162036 AG +/-
MTRR R415T rs2287780 CC -/-
MTRR A664A rs1802059 GG -/-
BHMT-02 rs567754 TT +/+
BHMT-04 rs617219 CC +/+
BHMT-08 rs651852 TT +/+
AHCY-01 rs819147 TT -/-
AHCY-02 rs819134 AA -/-
AHCY-19 rs819171 TT -/-
CBS C699T rs234706 AG +/-
CBS A360A rs1801181 AG +/-
CBS N212N rs2298758 GG -/-
SHMT1 C1420T rs1979277 AA +/+
Yea the raw data is what I’ve been using thus far. There are a couple little programs I wrote. Another one will search a group of genomes for a certain gene - which is probably the same thing as what @awor has been doing with excel?
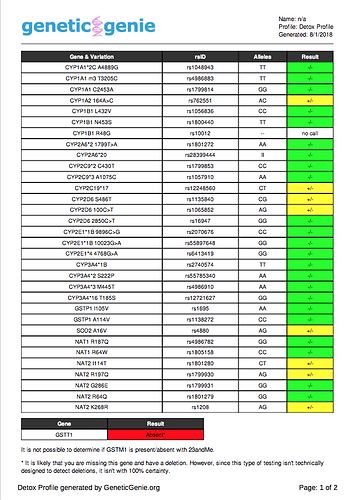
Here are my Methylation and Detox results from genetic genie using the 23andme raw data.
MTHFR Mutations
Heterozygous (+/-):
MTHFR C677T
Homozygous (+/+)
VDR Taq
MAO-A R297R
Heterozygous (+/-)
MTR A2756G
MTRR A66G
MTRR A664A
AHCY-01
AHCY-02
AHCY-19
CBS A360A
SHMT1 C1420T
Detox Profile Results
GSTT1 is Absent
My current focus is comparing allele frequency/distribution of patients vs. reference databases in dbsnp. It will get interesting once (and if) we find genes which have a very different distribution in our cohort vs the dbsnp control populations. I will publish this data here once I am finished.
Awor, you should contact Droit. We had an extensive list at one point looking at all of these data points for methylation and detox pathways.
I got the list thx moonman - it was in a spreadsheet on solve (unless this is a different one)
We had this one saved on OneDrive. Can you post the one you found, I know I had a number of different spreadsheets.
I’ve sent you a private msg
Hey guys, an update on this project. @awor and myself just had a great call with @Ghost and I’m really pleased we’ll be working on this together.
Please request and download your raw data from 23andme.
This comes in the form of a text file around 15 megabytes, and shortly we’ll be devising a way for you to get this to us. There’s a button in your 23andme account to request this. Anyone who hasn’t yet and wants to do a 23andme is welcome to do so (if they can afford it) to contribute to this dataset.
Thanks