Hi - I did 23andMe and would be happy to share my results.
Welcome to Propeciahelp Ghost! Glad to have you here.
That would be great, thanks for your offer. For the moment and for what I am doing, it would serve me well if you could just give me your GeneticGenie report, similar as to others who have posted in the past. You can either pm me or post it here, whatever you prefer. I am then hoping to look at it together with @Ghost, he might be interested in raw data by the sound of it. You can always give us that later, if required.
Gene & Variation rsID Alleles Result
COMT V158M rs4680 AG +/-
COMT H62H rs4633 CT +/-
COMT P199P rs769224 GG -/-
VDR Bsm rs1544410 CC -/-
VDR Taq rs731236 AG +/-
MAO A R297R rs6323 T +/+
ACAT1-02 rs3741049 GG -/-
MTHFR C677T rs1801133 GG -/-
MTHFR 03 P39P rs2066470 AG +/-
MTHFR A1298C rs1801131 GT +/-
MTR A2756G rs1805087 AG +/-
MTRR A66G rs1801394 AG +/-
MTRR H595Y rs10380 CT +/-
MTRR K350A rs162036 AG +/-
MTRR R415T rs2287780 CC -/-
MTRR A664A rs1802059 GG -/-
BHMT-02 rs567754 TT +/+
BHMT-04 rs617219 CC +/+
BHMT-08 rs651852 TT +/+
AHCY-01 rs819147 TT -/-
AHCY-02 rs819134 AA -/-
AHCY-19 rs819171 TT -/-
CBS C699T rs234706 AG +/-
CBS A360A rs1801181 AG +/-
CBS N212N rs2298758 GG -/-
SHMT1 C1420T rs1979277 AA +/+
Yea the raw data is what I’ve been using thus far. There are a couple little programs I wrote. Another one will search a group of genomes for a certain gene - which is probably the same thing as what @awor has been doing with excel?
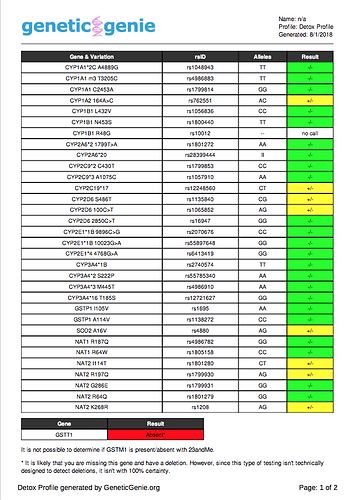
Here are my Methylation and Detox results from genetic genie using the 23andme raw data.
MTHFR Mutations
Heterozygous (+/-):
MTHFR C677T
Homozygous (+/+)
VDR Taq
MAO-A R297R
Heterozygous (+/-)
MTR A2756G
MTRR A66G
MTRR A664A
AHCY-01
AHCY-02
AHCY-19
CBS A360A
SHMT1 C1420T
Detox Profile Results
GSTT1 is Absent
My current focus is comparing allele frequency/distribution of patients vs. reference databases in dbsnp. It will get interesting once (and if) we find genes which have a very different distribution in our cohort vs the dbsnp control populations. I will publish this data here once I am finished.
Awor, you should contact Droit. We had an extensive list at one point looking at all of these data points for methylation and detox pathways.
I got the list thx moonman - it was in a spreadsheet on solve (unless this is a different one)
We had this one saved on OneDrive. Can you post the one you found, I know I had a number of different spreadsheets.
I’ve sent you a private msg
Hey guys, an update on this project. @awor and myself just had a great call with @Ghost and I’m really pleased we’ll be working on this together.
Please request and download your raw data from 23andme.
This comes in the form of a text file around 15 megabytes, and shortly we’ll be devising a way for you to get this to us. There’s a button in your 23andme account to request this. Anyone who hasn’t yet and wants to do a 23andme is welcome to do so (if they can afford it) to contribute to this dataset.
Thanks
Added 23AndMe to the community campaigns page.
Have ordered one myself and will send the results over.
Let’s keep this thing rolling guys 
Anyone who orders one - you’re a champ!
So it is the 23 and me plus health test we need to order? They have one that is 69 and the health 139$ now I believe…
I don’t know if these 23andMe test results are accurate you guys. Might be a waste of time and a money grab from the companies doing them.
If you live in the US, you get the whole test. Outside the US, you get only the smaller one (just files). This files can be analysed through different online platforms to be found in this thread. Best
At least in the PSSD research side of things, we’re only interested in the .txt raw data. Paying the extra money gets you the exact same data recorded, but you also have access to a lot of 23andMe’s proprietary research that will tell you anything from how tall you should be to if you are likely to go bald. For me, those things aren’t what we’re looking for, and are maybe a waste of money. It’s usually those trait facts that are incorrect and not the genotyping itself.
Promethease offers a health report that is perhaps even better than the one 23andMe offered (as of 2014) and it only costs $12.
Never found much evidence of 23andMe being erroneous, and believe me, I went looking for information on error rates after this: 23andMe Test and Cystic Fibrosis Gene
All sorts of different sources quote different error rates, but it appears to fall between 0.01% and %1.0, in the material I have seen, with a vast majority of the references citing numbers toward the lower end, between 0.01% and 0.05%. Most of the high error rates were attributed to poor handling of DNA samples or a just-plain-bunk probe for some of the SNPs. Pretty reliable for a ~$100 test, if you ask me.
In fact, the SNP genotyping they do is generally considered more accurate than whole-genome sequencing alone, which costs ~$1000. If ever considering whole-exome sequencing, save your money for something better.
Hey Awor,
i know, this is very speculative, but do you think, that IF (a very very big IF) the MTRR A66G Gene is the root cause of our problem, it would potentially be solveable with CRISPR?
I have 23andme Data as well and will provide.
I’m aware of the conflicting information over the accuracy of DTC tests, but it’s accurate enough for the programmatic correlation we’re interested in doing. We believe it’s worthwhile for those with the data to share it, and if any more members want to have it done to contribute more data then that’s great.
As @Ghost says we’re interested in the raw .txt file and will post a way for users to get that to us soon, so if previous posters can grab it in the meantime that would be helpful.
There is precedent for such data being used in attempts to predict drug responses: