I’m doubting the efficacy of this injecting method but will stick with it until I get ahold of Shippen Monday. Also, Awor is still tweaking his treatments and is going in for another round soon so alot of work yet to be done. Alot of eyes on this now. This idea has really got the interest of some of the PFS docs.
en.wikipedia.org/wiki/DNA_methylation
Detection
DNA methylation can be detected by the following assays currently used in scientific research:
* Methylation-Specific PCR (MSP), which is based on a chemical reaction of sodium bisulfite with DNA that converts unmethylated cytosines of CpG dinucleotides to uracil or UpG, followed by traditional PCR. However, methylated cytosines will not be converted in this process, and primers are designed to overlap the CpG site of interest, which allows one to determine methylation status as methylated or unmethylated.
* The HELP assay, which is based on restriction enzymes' differential ability to recognize and cleave methylated and unmethylated CpG DNA sites.
* ChIP-on-chip assays, which is based on the ability of commercially prepared antibodies to bind to DNA methylation-associated proteins like MCP2.
* Restriction landmark genomic scanning, a complicated and now rarely-used assay based upon restriction enzymes' differential recognition of methylated and unmethylated CpG sites; the assay is similar in concept to the HELP assay.
* Methylated DNA immunoprecipitation (MeDIP), analogous to chromatin immunoprecipitation, immunoprecipitation is used to isolate methylated DNA fragments for input into DNA detection methods such as DNA microarrays (MeDIP-chip) or DNA sequencing (MeDIP-seq).
* Molecular break light assay for DNA adenine methyltransferase activity – an assay that relies on the specificity of the restriction enzyme DpnI for fully methylated (adenine methylation) GATC sites in an oligonucleotide labeled with a fluorophore and quencher. The adenine methyltransferase methylates the oligonucleotide making it a substrate for DpnI. Cutting of the oligonucleotide by DpnI gives rise to a fluorescence increase.
Software
Free
* Many software packages are available to analyse bioinformatic data, many of them free.
Commercial
* Qlucore Omics Explorer among others (offers PCA and hierarchical clustering)
en.wikipedia.org/wiki/Human_Epigenome_Project
Human Epigenome Project
From Wikipedia, the free encyclopedia
Jump to: navigation, search
Human Epigenome Project (HEP) is a multinational science project, with the stated aim to “identify, catalog, and interpret genome-wide DNA methylation patterns of all human genes in all major tissues”. It is financed by government funds as well as private investment, via a consortium of genetic research organizations.
The call for such a project was widely suggested a supported by cancer research scientists from all over the world. Consortium
The HEP consortium is made up of the following organizations:
* The Wellcome Trust Sanger Institute — UK
* Epigenomics AG — Germany/USA
* The Centre National de Génotypage — Francecell.com/retrieve/pii/S0092867406003850
Article
Add/View Comments (0)
JHDM2A, a JmjC-Containing H3K9 Demethylase, Facilitates Transcription Activation by Androgen Receptor
Kenichi Yamane1, 2, Charalambos Toumazou3, Yu-ichi Tsukada1, 2, Hediye Erdjument-Bromage4, Paul Tempst4, Jiemin Wong3 and Yi Zhang1, 2, Go To Corresponding Author,
1 Howard Hughes Medical Institute, Lineberger Comprehensive Cancer Center, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599, USA
2 Department of Biochemistry and Biophysics, Lineberger Comprehensive Cancer Center, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599, USA
3 Department of Molecular and Cellular Biology, Baylor College of Medicine, One Baylor Plaza, Houston, TX 77030, USA
4 Molecular Biology Program, Memorial Sloan Kettering Cancer Center, 1275 York Avenue, New York, NY 10021, USA
Ph: 919-843-8225; Fax: 919-966-4330
*
Summary
* Covalent modification of histones plays an important role in regulating chromatin dynamics and transcription. Histone methylation was thought to be an irreversible modification until recently. Using a biochemical assay coupled with chromatography, we have purified a JmjC domain-containing protein, JHDM2A, which specifically demethylates mono- and dimethyl-H3K9. Similar to JHDM1, JHDM2A-mediated histone demethylation requires cofactors Fe(II) and α-ketoglutarate. Mutational studies indicate that a JmjC domain and a zinc finger present in JHDM2A are required for its enzymatic activity. Overexpression of JHDM2A greatly reduced the H3K9 methylation level in vivo. Knockdown of JHDM2A results in an increase in the dimethyl-K9 levels at the promoter region of a subset of genes concomitant with decrease in their expression. Finally, JHDM2A exhibits hormone-dependent recruitment to androgen-receptor target genes, resulting in H3K9 demethylation and transcriptional activation. Thus, our work identifies a histone demethylase and links its function to hormone-dependent transcriptional activation.Read that. Question here is is there a way to target the androgen receptor gene for demethylation?
Actually thats a good question. I don’t think thats possible yet. Am i right in thinking that drugs like procaine or azacitidine have a generalised demethylating effect? So if you take too much it can cause problems?
If I understand Awor correctly the Procain IV treatment globally demethylates.I would guess there should be some research toward targeting genes as I would guess that is what they would attempt in cancer treatment. Awor is due for another treatment soon so will definately be interesting to see how it turns out. Seems very promising.
Found some useful studies on lysine-specific demethylase (LSD1) which i felt may be relevant to this topic.
I’ll also post this in the other studies section.
Cooperative demethylation by JMJD2C and LSD1 promotes androgen receptor-dependent gene expression.
Posttranslational modifications of histones, such as methylation, regulate chromatin structure and gene expression. Recently, lysine-specific demethylase 1 (LSD1), the first histone demethylase, was identified. LSD1 interacts with the androgen receptor and promotes androgen-dependent transcription of target genes by ligand-induced demethylation of mono- and dimethylated histone H3 at Lys 9 (H3K9) only.
ncbi.nlm.nih.gov/pubmed/17277772
LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription.
LSD1 relieves repressive histone marks by demethylation of histone H3 at lysine 9 (H3-K9), thereby leading to de-repression of androgen receptor target genes…Thus, modulation of LSD1 activity offers a new strategy to regulate androgen receptor functions. Here, we link demethylation of a repressive histone mark with androgen-receptor-dependent gene activation, thus providing a mechanism by which demethylases control specific gene expression.
Merck said that Propecia would simply stop the bad type of testosterone and allow us to regain hair. If we experienced some of the few possible side effects they would go away upon discontinuation of the drug.
We have only a partial handle on how PFS sufferers are affected and treatments that work initially but diminish in returns over time. Gene demethylating is a new subject and we have some of the first people in the world experimenting with such treatments trying to cure something based on symptoms because there are no lab tests.
This is not simple and anyone who thinks it is is kidding themselves.
I think Awor is definately onto something here. As his treatment continues should tell us alot. This might be an incorrect anaology, but sticking to the facts we know - We are all vit d deficient, some even after taking high supplements. Since vit d acts like a hormone wouldnt this insensitivity theory explain that?
ps- Just spoke to Shippens office and he wants me to continue w/Procaine even though Awor is saying sub q injections will have no effect. I’m really confused now. 
pps- I’ve read some anechdotal reports that Procaine treatments can reverse alopecia. Isnt that ironic?
Another interesting group of studies specifically related to 5 alpha reductase.
I’ll post this in the other studies section.
Evidence that steroid 5alpha-reductase isozyme genes are differentially methylated in human lymphocytes.
The overall results suggest that 5alpha-reductase genes 1 and 2 are differentially methylated in lymphocytes from normal and 5alpha-reductase deficient patients. Moreover, the extensive cytosine methylation pattern observed in exon 4 of 5alpha-reductase 2 gene in deficient patients, points out to an increased rate of mutations in this gene.
ncbi.nlm.nih.gov/pubmed/11948017
Differential methylation in steroid 5 alpha-reductase isozyme genes in epididymis, testis, and liver of the adult rat.
Our results suggest that the differential methylation pattern in 5 alpha-reductase gene 2 in reproductive and nonreproductive tissues should be involved in the regulation of its expression.
This could be very important, IF methylation is the problem.
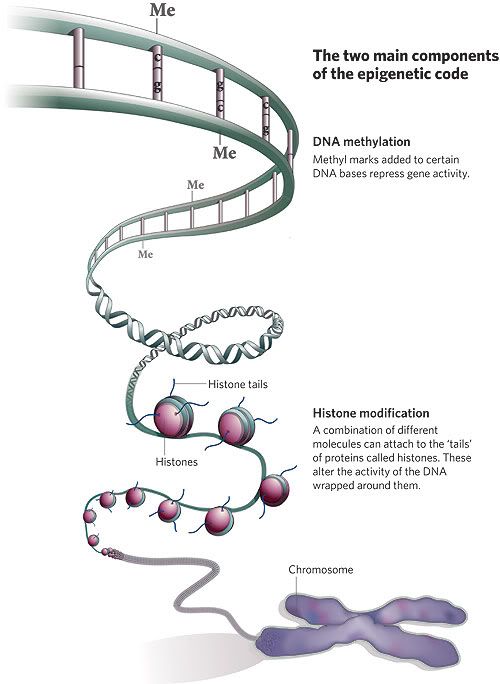
Awor mentions this paper[1] to support his use of procaine to demethylate, however it doesnt actually mention androgen receptors. It mentions procaine to demethylate where there is an addition of methyl groups to the DNA, mostly at CpG sites, promotor regions.(in cancer)
However it appears there can be in two types of Methylation; DNA Methylation or Protein (post-translational) Methylation [2][3], where histones are modified by adding methyl groups thus changing gene expression.
LSD1 (and JMJD2C[5]) actually works using the demethylation of histones. Which is this second Protein/post translational form of methylation. “LSD1 relieves repressive histone marks by demethylation of histone H3 at lysine 9 (H3-K9), thereby leading to de-repression of androgen receptor target genes”[4]
Therefore both seem like valid ways to demethylate ARs. Prehaps those enzymes more so if histone methylation is in fact one of the normal/natural ways to AR expression is controlled? LSD1/JMJD2C also appear more directed to specifically demethylate ARs, which would appear to be safer.
But LSD1/JMJD2C should occur naturally in our bodies. Do we therefore need to purchase some from a lab and inject ourselves with it because higher serum levels will cause it to ‘work’ more? I guess theres only one way to find out.
[1] cancerres.aacrjournals.org/content/63/16/4984.full
[2] en.wikipedia.org/wiki/Methylation#Epigenetics
[3] en.wikipedia.org/wiki/Epigenetic_inheritance#DNA_methylation_and_chromatin_remodeling
[4] nature.com/nature/journal/v437/n7057/abs/nature04020.html
[5] ncbi.nlm.nih.gov/pubmed/17277772
OK, bit of a shot in the dark here…
As per the studies posted above, one method of de-methylation appears to be through demethylases such as lysine specific demethylase.
Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases.
It has been unclear whether demethylases exist that reverse lysine trimethylation. We show the JmjC domain-containing protein JMJD2A reversed trimethylated H3-K9/K36 to di- but not mono- or unmethylated products.
ncbi.nlm.nih.gov/pubmed/16603238
I came across some information regarding this protein, or family of proteins, most interestingly that its activity, and thus theoretically its ability to de-methylate, could be upregulated by alpha-ketoglutaric acid.
alpha-ketoglutaric acid
[Iron co-treated with alpha-ketoglutaric acid] results in increased activity of JMJD1A protein] which results in decreased methylation of HIST3H3 protein
genecards.org/cgi-bin/carddisp.pl?gene=KDM3A
α-Ketoglutaric acid is sold as a dietary supplement and to body builders as AKG or a-KG. Some believe it increases stamina.
en.wikipedia.org/wiki/Alpha-Ketoglutaric_acid
So, yeah as i said this is a long shot but fortunately one with no downside risk as it appears to be a fairly safe supplement.
Polyphenols contained in coffee beans have shown de-methylating properties.
Inhibition of DNA methylation by caffeic acid and chlorogenic acid, two common catechol-containing coffee polyphenols.
…we found that the presence of caffeic acid or chlorogenic acid inhibited in a concentration-dependent manner the DNA methylation catalyzed by prokaryotic M.SssI DNA methyltransferase (DNMT) and human DNMT1…The maximal in vitro inhibition of DNA methylation was approximately 80% when the highest concentration (20 microM) of caffeic acid or chlorogenic acid was tested…The presence of caffeic acid or chlorogenic acid inhibited DNA methylation predominantly through a non-competitive mechanism, and this inhibition was largely due to the increased formation of S-adenosyl-L-homocysteine (SAH, a potent inhibitor of DNA methylation), resulting from the catechol-O-methyltransferase (COMT)-mediated O-methylation of these dietary catechols.
ncbi.nlm.nih.gov/pubmed/16081510
The easiest way to consume this is probably a green coffee bean extract.
Also, as mentioned previously in this thread, green tea extract seems to share similar de-methylating properties.
Reversal of hypermethylation and reactivation of genes by dietary polyphenolic compounds
ncbi.nlm.nih.gov/pmc/articles/PMC2829855/
They observed that EGCG directly inhibited DNMT activity and partially reversed RAR-β methylation status. Other catechol polyphenols inhibited DNMT indirectly by being methylated and converting S-adenosyl-L-methionine (SAM) to S-adenosly-L-homocysteine (SAH), which is a strong inhibitor of DNMT. Caffeic acid and chlorogenic acid also partially inhibited methylation of the promoter region of the RAR-β gene in breast cancer cell lines.4
RG108. Appears to be a DNA methyltransferase (DNMT) inhibitor developed specifically for the purpose of reactivating silenced tumor suppressor genes.
Abstract
DNA methylation regulates gene expression in normal and malignant cells. The possibility to reactivate epigenetically silenced genes has generated considerable interest in the development of DNA methyltransferase inhibitors. Here, we provide a detailed characterization of RG108, a novel small molecule that effectively blocked DNA methyltransferases in vitro and did not cause covalent enzyme trapping in human cell lines. Incubation of cells with low micromolar concentrations of the compound resulted in significant demethylation of genomic DNA without any detectable toxicity. Intriguingly, RG108 caused demethylation and reactivation of tumor suppressor genes, but it did not affect the methylation of centromeric satellite sequences. These results establish RG108 as a DNA methyltransferase inhibitor with fundamentally novel characteristics that will be particularly useful for the experimental modulation of epigenetic gene regulation.
cancerres.aacrjournals.org/content/65/14/6305.full#abstract-1
Psammaplin A, found in a marine sponge, is also a potential DNA demethylator.
Psammaplins from the sponge Pseudoceratina purpurea: inhibition of both histone deacetylase and DNA methyltransferase.
The structures of psammaplins E (9) and F (10), which each contain an oxalyl group rarely found in marine organisms, were determined by spectroscopic analysis. Compounds 4, 5, and 10 are potent histone deacetylase inhibitors and also show mild cytotoxicity. Furthermore, compounds 4, 5, and 11 are potent DNA methyltransferase inhibitors.
Previously mentioned in this thread was Sodium Butyrate, an HDAC inhibitor. This article gives some insight into the fact that HDAC inhibitors may not be necessarily useful on their own but could be useful in conjunction with a DNA demethyltransferase inhibitor.
nature.com/nrclinonc/journal/v2/n12s/full/ncponc0346.html#B9
ncbi.nlm.nih.gov/pubmed/19501473 Epigenetic side-effects of common pharmaceuticals: a potential new field in medicine and pharmacology
Great finds! How we can put these thoughts into action?
I’m afraid we can’t…yet.