Even if Khera was to determine which genes have been silenced, at this stage there’d be little we could do about it anyhow.
Collagen synthesis is dependent on sex hormones although not sure which ones.
Hopefully it can return to normal or at least near normal if those hormones are restored. I expect some skin damage to persist like the deep stretch marks untill technology gets better, but even if it was firm again I’d be happy. Look like a pale ghost now lol but it could be worse I guess.
- The study shows that out of 16 PFS patients, close to 60% of them had issues with SRD5A2 - the gene that allows normal functioning of steroid synthesis in the body.
- Out of the controls studied (people who did not take PFS), only 7% showed problems with SRD5A2.
A few anecdotal conclusions can be drawn:
- Most PFS’ers have issues creating steroids due to alterations in the gene SRD5A2 (yes only 16 were studied, but there are not many people with PFS so when you consider that it’s actually a decent sample size percentage-wise)
- Whatever causes these issues with SRD5A2 can be caused by variables other than Finasteride since the same issue was seen in at least one of the non-Finasteride users.
*edit 7/22/2019 - After reading the full study I’ve changed my opinion - 100% of the non-pfs SRD5A2 controls had UN-Methylated SRD5A2 (normal) levels (there was one who had methylated SRD5A2 but he suffered from normotensive hydrocephalus (a condition which has symptoms like difficulty concentrating and urinary issues) and should have been excluded from the “healthy” controls.
I was looking for a stronger correlation and after seeing the fully study this looks to be a very strong correlation.
Yes, a few things, but mainly cortisol.
Yes, as with every other symptom, site specific over-expression of the AR, which importantly is already established in 100% of symptomatic dermal prepuce tissue across cell lines in PFS. The prevalence of the symptom you mention and this important primary finding already disproves the hypothesis of effects localised to the CNS.
Androgen signalling has a determinant role in skin maintenance, epithelialisation, collagen deposition and inflammation. AR signalling is inhibitory of skin healing. (1-7)
- Gilliver, Regulatory roles of androgens in cutaneous wound healing, 2002
- Ashcroft and Mills, Androgen receptor-mediated inhibition of cutaneous wound healing, 2002
- Gilliver et al, 5alpha-dihydrotestosterone (DHT) retards wound closure by inhibiting re-epithelialization, 2009
- Ashcroft et al, Topical estrogen accelerates cutaneous wound healing in aged humans associated with an altered inflammatory response, 1999
- Fimmel and Zouboulis, Influence of physiological androgen levels on wound healing and immune status in men, 2005
- Lai et al Monocyte/macrophage androgen receptor suppresses cutaneous wound healing in mice by enhancing local TNF-alpha expression, 2009
- Gilliver et al, Androgens influence expression of matrix proteins and proteolytic factors during cutaneous wound healing, 2007
…As such antiandrogenic substances are increasingly considered as potential therapeutic angles to aid healing - including finasteride, quercetin and flutamide (8, 9, 10):
- Toraldo et al, Topical androgen antagonism promotes cutaneous wound healing without systemic androgen deprivation by blocking β-catenin nuclear translocation and cross-talk with TGF-β signaling in keratinocytes, 2012
- Liu, Quercetin suppresses glomerulosclerosis and TGF‑β signaling in a rat model, 2019
- Wang et al, Androgen actions in mouse wound healing: Minimal in vivo effects of local antiandrogen delivery, 2016.
This is likely part of a pleiotropic picture as increased androgen signalling has been demonstrated to be inhibitory of healing following injury to androgen target tissues such as the heart and endometrium. Finasteride has recently been suggested as potentially therapeutic in heart failure due to marked improvement in animal models.
Hopefully I can clear this up, as this is obviously coming up a lot: What Melcangi has done is not on remotely the same scale. While more confirmation of epigenetic differences is very good to have (I gave a large personal donation to this particular study and was hopeful it could provide some fast evidence of methylation in PFS patients), 5ar deficiency is not a plausible driver of PFS as @awor has stated for many years, and this result should make clear. The practicality of this will lie in more evidence for a direction towards epigenetic involvement, again as @awor said.
This present study, at least in organisation terms, got underway at the end of 2017. It looked at two genes in serum and CSF. I think it’s therefore simple to understand that it is much more of an undertaking to look at those, plus 19998 more, in tissue biopsy of a larger control-matched cohort, with what we hope is the potential to provide insight into possible driving factors of previously established AR deregulation. We expect a variably significant picture of altered expression depending on the severity of case, and disentangling the volume of data is not easy.
I do not know the quality of report that will ultimately be produced or factors that may influence that. However, that a narrowly-focused study is quick does not make it “shameful” that the important things inherently take longer. It is not in the same ball park.
Does anyone know anything about Melcangi’s other ongoing PFS study, on the gut micriobiome, and how would we evaluate the significance of that?
i think melcangi is the only one that can help us. i have no hopes in baylor
@awor, @axolotl Do you guys think that the fact that not only all of the pfs sufferers showed srd5a2 hypermethylation proves that srd5a2 hypermethylation is not the root cause of the problem, and from now on it would be better to focus on androgen receptor’s expression?
Although is important to understand what the gene’s hypermethylation could imply because the difference is strongly statistically significant.
What I would expect is the more genes they look at a pattern like this should start emerging showing a variance of genes that are affected and at different percent in different patients…So a whole host of genes affected but at different degrees in different people could explain why some get crippled, some get fat, some lose weight, some recover more etc…Why yes, its likely they could not do anything right now I suppose about it but wouldn’t you finally like to know the cause and see this proven?? So people would quit coming up with all these outlandish theories…
I suppose that would certainly be one major benefit, ey? 
Khera said he is not to find a treatment but the
'cause" only…Thats his job is to find the cause…But the problem I had as moonchild said he just Started comparing them?! uhhhh
Thanks for the comprehensive reply @axolotl. It’s great to see all the pieces finally coming together with this condition.
It’s interesting that you mentioned antiandrogenic substances being a potential aid for wound healing, but for PFS patients that just causes us to crash further right?
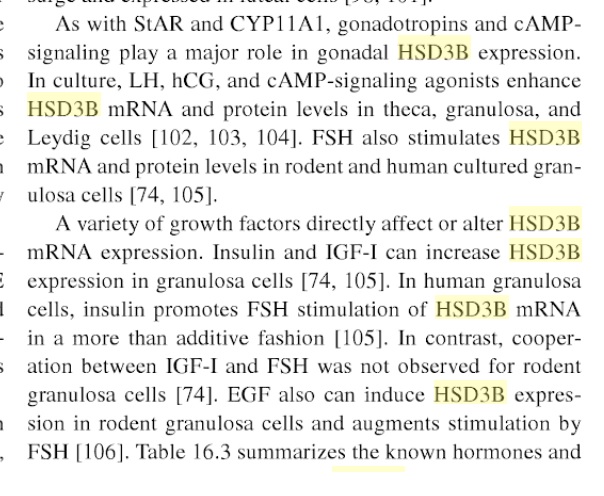
It can be tested i guess. And cyp11a1 might be one of the key issues.
And one thing sodium bicarbonate does…Increases cAMP signaling…
https://academic.oup.com/jcem/article/90/8/4858/2838533
Treatment with atRA significantly augmented CYP17, CYP11A1, and StAR mRNA accumulation with maximal induction occurring within 16–24 h after treatment (Fig. 3).
Other study shows different results ? or i dont undestand something
We also investigated the effects of atRA on the expression levels of StAR, Cyp11a1, and 3β-hydroxysteroid dehydrogenase (3β-HSD) mRNAs. GCs were cultured for 3–12 h with or without atRA (30 μM; Fig. 2B–D). Treatment with atRA resulted in significant augmentation of StAR
and Cyp11a1 mRNA levels at all times. In contrast, 3β-HSD mRNA was unaltered
Yep, different effects of RA on 3b-HSD in glial cells and granulosa cells from the looks of it. The study I was referencing:
-“ATRA also strongly induced the expression of steroidogenic acute regulatory protein (StAR) and 3beta-hydroxysteroid dehydrogenase (3beta-HSD) (an increase of 5- and 50-fold, respectively).”
I have read the study, very nice one! so if we inhibitthe methilation we can have normal neurosteriods and feel better?
I think its very strong correlation specially after the one i the control group that had it had some specific health issue (I dont know if its related with that gene). And also open the possibility to study the cause of the alteration using animal models and also give us the opportunity of a new protocol.
