Sent you a PM with upload instructions.
Thank you.

Sent you a PM with upload instructions.
Thank you.
The most simple form of association analysis of the 23andMe v5-chip data submitted has been completed. This only included autosomes (chromosomes 1-22). As was to be expected, nothing considered statistically significant was found. There was also not much in the way of clustering, which may indicate a genetic locus associated with a disease in question:
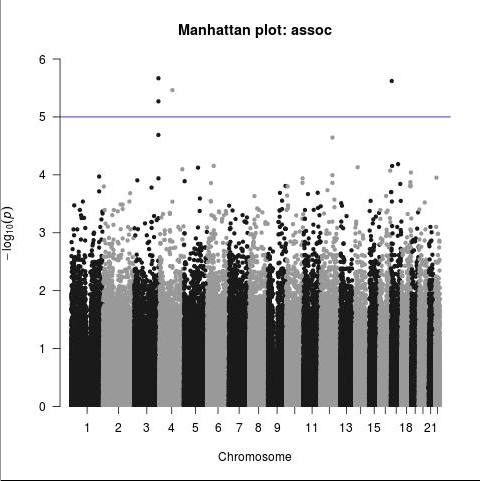
The only SNPs beyond the 1E-5 threshold (still not considered significant at the per-SNP level in GWAS) mapped to 2 genes, P3H2/LEPREL1 and WSCD1, which don’t appear likely to be involved in any mechanism of a post-drug syndrome; either hypothesized mechanisms, or wildly speculated mechanisms. It’s probably a “fluke.” Further analysis will strengthen or weaken this idea.
The file containing the results was submitted to a gene ontology service, which attempts to characterize GWAS data according to pathways, functions, or traits, that involve significantly-associated genes. In the absence of significant associations, the service still reports the top-10 pathways, functions, or traits, represented by the data. Basically, the lowest quality setting allowed had to be used for the data to be processed:
Keep in mind that these reported associations were extremely weak and that basing any treatments on something so flimsy is a waste of time and money and could be another risk to your health. Also keep in mind that this is an amateur GWAS study at this point and intended to find something worth approaching professionals with.
There was a bit of a change in plans and the next step will be imputation, which confidently infers the existence of unobserved SNPs based off of observed SNPs in an individual’s genome. Basically, this will provide much more data to work with. Imputation is a computationally intensive process in GWAS, so it may take some time. I’m also committed to some Accutane things in the coming weeks and may not have much time to devote to propeciahelp or this project.
Thanks for the update.
What was the motivation behind the change in plans?
Appreciate the update Dubya, posts are always very informative. It really spells out how complicated this is.
For one, our geneticist friend has expressed his assessment that imputation is needed to sufficiently increase the power of a study with so few genomes. He has given no indication that he is anything but a professional in his field and, as an amateur, I am compelled to give his judgement some weight. I think it’s better to just do the imputation, then analysis, rather than waste time with analysis, imputation, re-analysis.
Also, a forum member who is acquainted with the founder of an imputation service put us in contact with him and he offered to help.
23andMe please.
Nothing against Ancestry, it’s just that our existing data is 23andMe and the 2 companies have used different tests with different chips that included different snps. We chose 23andMe primarily because many members were already genotyped through their service.
Thanks Dubya for the hard work!
1.) Can you explain a little bit more about the top-10 pathways? They are non-significant but show a correlation?
If I see it right “chromatin remodeling” shows the highest correlation?
Though not significant that could be something, not?
There is a connection to epigenetic regulation, senescence and the CHARGE-syndrome that include “micropenis” and “delayed of absent pubertal development”.
If not too much work, it would be nice to see the SNP-name of the top SNPs (to look at them in SNPedia).
2.) Does the data set include the SNPs of the whole 5 isoenzymes of the 5ar-family as well as the SNP of the androgene receptor?:
5α-R1,
5α-R2,
5α-R3,
GPSN2,
GPSN2L
Thanks!
Just ran across this post. Have my 23 and me raw data ready for up load. Am I able to submit?
@Akiyah, Yes. I will PM you.
@anon5006275, I’ll check again to see if the list of snps associated with chromatin remodeling are accessible. I tried briefly once already. Remember that the association is extremely weak, even for a peripheral gene set.
Also, I’m pretty certain the 5-arI, 5-arII, AR genes, and their flanking regions have already been sequenced in PFS patients in the studies and nothing unusual was found.
In what way are you interested in GPSN2? It is one of the genes whose transcription was repressed ~1.4 fold in a study of isotretinoin (Accutane) treated patients.
@Dubya_B can you check ApoE4 frequency among us ? It gives worst out comes with any kind of brain injury. It is also main risk factor for Late onset Alzheimer’s.
Also HTR2A snp’s might worth checking it is closely related pfc dopamine modulation, memory, pain perception and arousal.
The ‘T’ allele has been associated with more receptors/increased gene expression and more active receptors. The ‘ TT’ genotype has been associated with depression, panic disorder, and higher stress response. [71, 72].
The ‘ T’ allele was associated with significant reductions in general health, vitality, and social function [73]. The ‘ T’ allele has also been associated with irritable bowel syndrome (IBS) [44] and chronic fatigue syndrome (CFS) [73].
The ‘ C’ allele is associated with an extraverted personality, Rheumatoid Arthritis and novelty seeking. ‘ CC’ = 3.6x increased risk of sexual dysfunction when taking SSRI Antidepressants. This SNP correlates with rs6313, as in the T allele here will result in the A allele by rs6313 [73].
Oh, wow! GPNS2 is an enzyme with the same function as 5ar. It belongs to the 5ar-family. As they have same structure and functionality it is very likely that also Finasteride will bind to it. The link to Accutane is highly interesting!
Ideally the data set includes also the genes for 5ar-3 - Prof. Zitzmann thinks that is the reason for PFS as he says it is more located in the brain.
GPSN2L should be looked at as well as it also has the same function and structure as 5ar. It belongs to the same isoenzyme family.
Any 23andMe Patient Data Analysis participant who wishes to access the summary results of their imputed genome, please send me a PM and you will be provided with a unique ID and instructions as soon as your data is complete.
The more recent (version-5) chip data is complete as of Feb 27th, with the exception of 1 or 2 genomes that seem to be hung up. The remainder should be available in the next few days.
Hello,
I just received my results from 23andme and I downloaded the raw data, how do I send it to you ?
“Acceptance of new submissions is closed until further notice and basic analysis on the current data set has begun. See the following post in this topic for more details:” oh shit … ok, too bad
Really cutting it close. Will send you a PM so you can supply it via email.
Thanks!
23andMe data can still be supplied by members who have an existing file; however, no one is actively encouraged to go through the trouble of having the test performed for the sake of this GWAS at this point.
Once the imputation has finished, and the second round of analysis has actually begun, it will be too late. There’s no set date of when this will be.
A “theory neutral” analysis of the 23andMe data of chromosomes 1-22 has been completed .
That is, no particular marker was intentionally searched for with bias, but a very “by the book” associative analysis of a subset of quality-controlled genotyped and imputed SNPs from chromosomes 1-22 was performed to see if any broke past the established ‘standard’ GWAS significance threshold of p<5x10^-8.
Unfortunately, and quite expectedly, no single SNP was observed beyond this significance threshold.
There didn’t appear to be any greater correlation between either case group or control group and over-represented SNPs in any trait/disease featured on ImputeMe.
Nothing in the top results of the gene or pathway analyses this round sticks out as indicative of epigenetic mechanisms, known diseases, common symptoms in PFS/PAS/PSSD, steroid metabolism, AR signalling, known diseases, immunity, autoimmunity, gluten intolerance, etc…
Basically, a proverbial “needle in the haystack” wasn’t discovered in these results.
However; there were multiple SNPs in a small region of chromosome 4 in the imputed data (only 1 of these was present in the genotyped 23andMe data) that were near the significance threshold at Nx10^-7. I am going to approach an acquaintance with GWAS experience to ask if this may be suggestive of anything. It appears to be an intergenic region of little known consequence.
Links to the summary results and gene and pathway analyses will be posted soon. Be patient.
If it is advisable, some more imputed genomes could be easily added to the analysis since the process is practically automated after all this hard work. A strictly post-finasteride patient analysis could also be easily performed soon. Chromosome X analysis may or may not be worth the time and effort but I will briefly look into it.
Some preliminary results:
Quality-controlled independent SNPs taken from genotypes imputed against a 1kGv3 reference panel by the Impute.Me service were used for the associative analysis. All pre and post-Imputation genomes were mapped to GRCh37
Selection of controls from 23andMe data obtained from OpenSNP was performed using the R package, PCAmatchR, prior to imputation.
QC and estimation of superpopulation was conducted according to 23andMe chip version, then intersecting SNPs across all 3 chip versions used were merged for the last 2 analyses.
SNPs on CHR 1-22 of All European samples genotyped on “v5” 23andMe.
Summary statistics: https://www.dropbox.com/s/1qj7o3o0qtsresg/sorted-v5Eur-assoc2.assoc?dl=0
FUMA results: https://fuma.ctglab.nl/browse/129
SNPs on CHR 1-22 of All European samples genotyped on v3, v4, and v5 23andMe chips
Summary statistics: https://www.dropbox.com/s/oz2ey7pt37tq2sf/sorted-vAllEur-assoc2.assoc?dl=0
FUMA results: https://fuma.ctglab.nl/browse/131
SNPs on CHR 1-22 of All samples genotyped on v3, v4, and v5 23andMe chips
Summary statistics: https://www.dropbox.com/s/9ledhm9bdh7bary/sorted-All-assoc2.assoc?dl=0
FUMA results: https://fuma.ctglab.nl/browse/130
The only “hotspot” (near significance threshold) was the chr4 ~10179769-10194628 region.
The only nearby genes were characterized as a couple pseudogenes, along with WDR1 and a non-coding micro-RNA, MIR3138, which appears to be located in an intron of WDR1.
Another analysis of the same data may be posted in the near future.
Even though the official submission period is over and we are no longer actively requesting 23andMe data, if anyone has recent 23andMe data they have obtained for personal use that they would like to contribute, please PM me. This analysis could be performed periodically for every ~10 or so genomes to add to the statistical power.
Thanks for the update, Dubya.
When you say preliminary results, what beyond what has already been analysed could occur at a later stage (aside from more test submission)?
It was mentioned earlier in this topic that someone with prior GWAS experience has helped a great deal as a guide with the analysis. Only about %5 (1.8 million / 36 million) of the SNPs from the imputed genomes passed the quality control checks and contributed to what was presented as preliminary results. I am hopeful that this person can help increase that number in a future analysis, without appreciably sacrificing the quality of the markers or the statistics derived from them. They have also made some good suggestions to improve interpretation of the results.