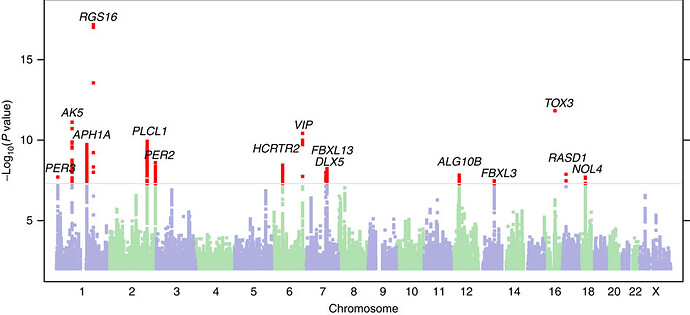
We currently have 33 genomes from mainly PFS patients, and another 14 from PSSD patients. I will attempt a first run at around 50 PFS genomes. As we all know, PFS is a very variable disease, both in scope and intensity. As written earlier, PFS is what is called a variable trait disease. From a genetics point of view, this indicates that a number of genes are likely involved. The more genes that are involved, the more samples we will need to cut through the statistical noise. The following picture shows a “Manhattan Plot”, which is a common way of representing GWAS results:
One can clearly see the noise I am talking about in this sample. Incidentally, this was from a study which looked at 1.1 million cases. I am not suggesting we need a million cases, but I hope that this example allows one to appreciated that even with 50 genomes, we will need a lot of luck to find something useful. It may be that we’ll need at least a few hundred genomes before we start to see something.
This work won’t produce an instant cure, but it will for sure contribute to better understanding our disease and hopefully provide some direction for future investigation. One of the key things we need to achieve is an animal model of PFS (rat, mouse). That will only be possible if we understand the genetic drivers of PFS, i.e. what makes US different from those many other guys, who apparently can take these substances without a problem. If we know those drivers, it should be possible to genetically engineer an animal so it develops PFS. Ideally, we would want it to get PFS quickly, and not have to wait 5 years. That means, we need to figure out which genes predict a quick onset of strong PFS, which further limits the available genomes.
Notice that I interchangeably am talking about genes, even though the data we have are snp’s. These represent locations on our chromosomes. Those locations will help find the underlying genes, but also here additional work is involved. Please appreciate that these kind of studies typically are run by a team of full time scientists. I am not a scientist, and doing this in my spare time. I have a rough understanding of what I am doing, but may run into limitations due to time and/or knowledge. If I reach that point, I am intending to get scientists involved to help. This will likely require further funding. If you are interested in this work and it’s objectives, you may want to save your donation money for this. If we get to the point where we need help, the funding will be collected through the Foundation, specifically ear marked for this study.